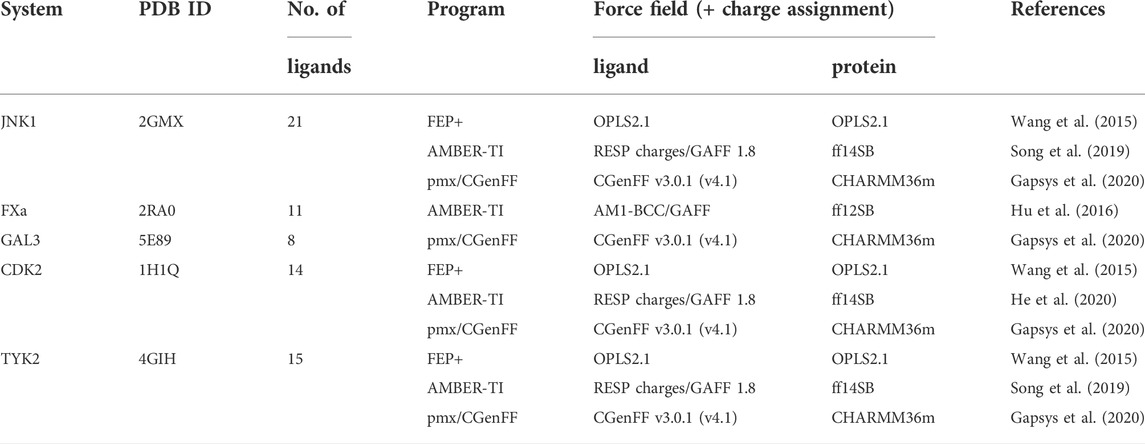
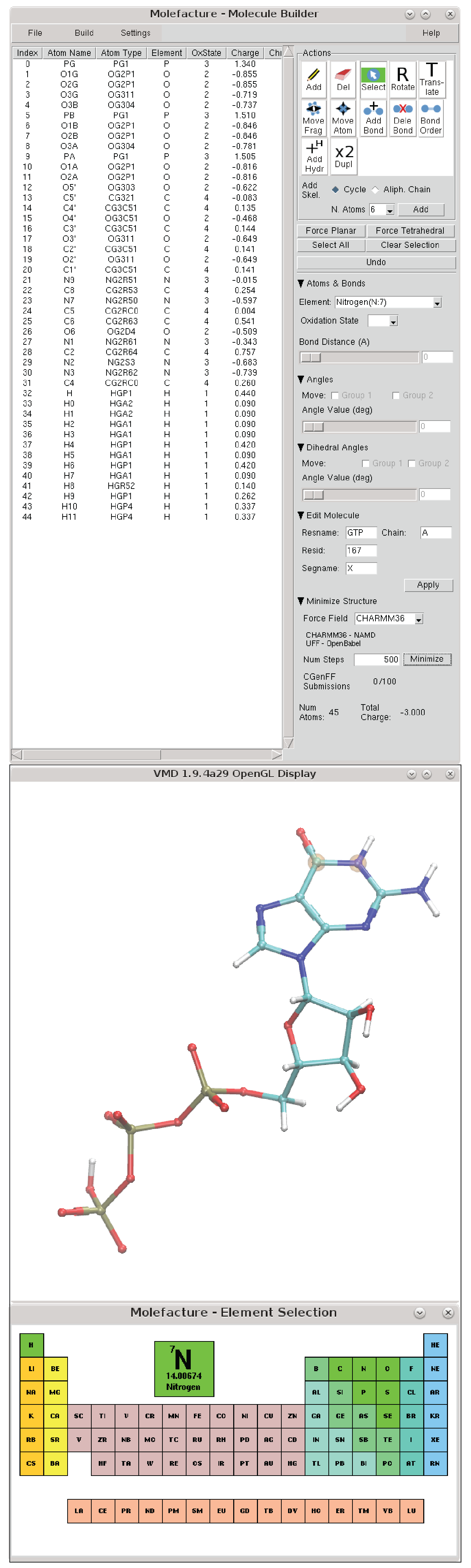
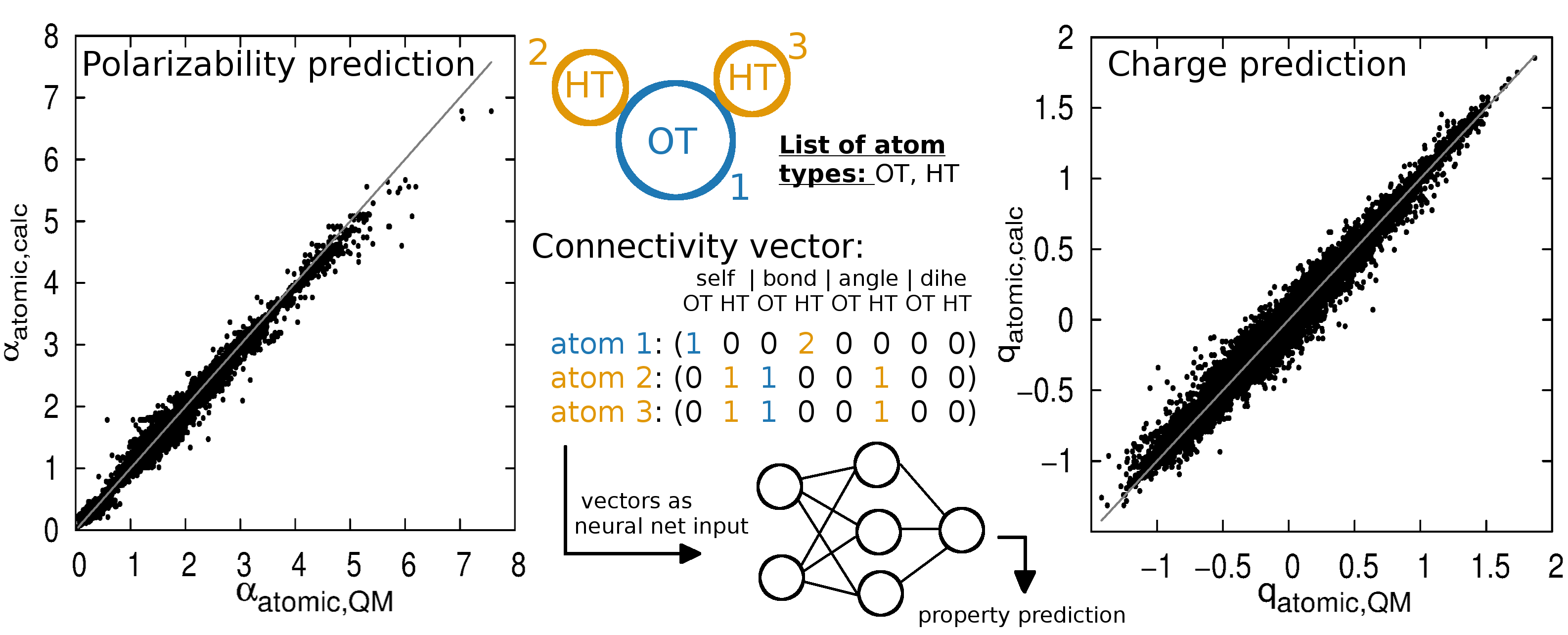
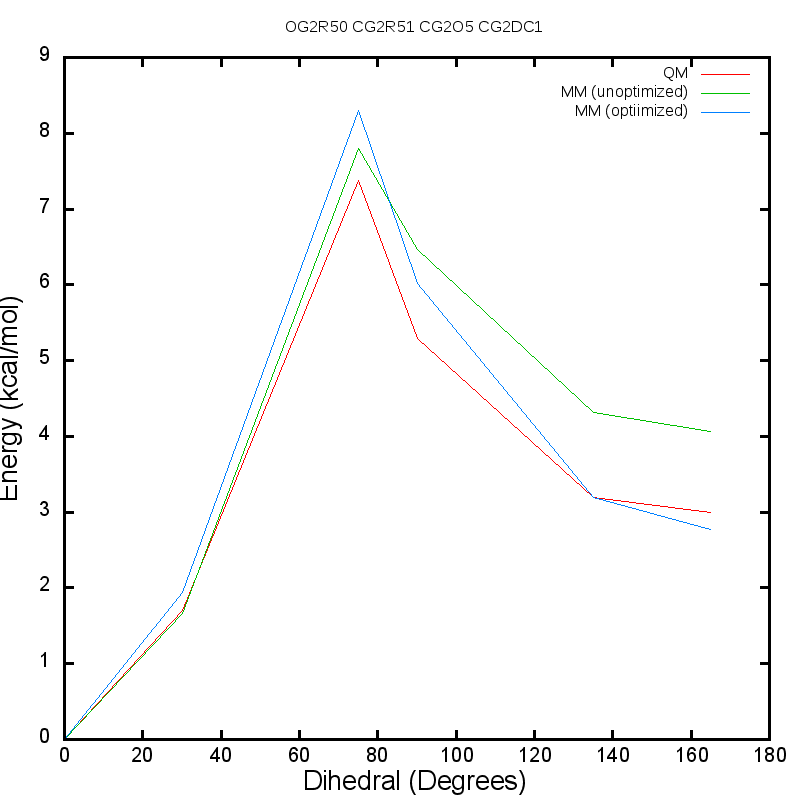
Additive CHARMM force field for naturally occurring modified ribonucleotides - Xu - 2016 - Journal of Computational Chemistry - Wiley Online Library

Automation of the CHARMM General Force Field (CGenFF) I: Bond Perception and Atom Typing | Semantic Scholar
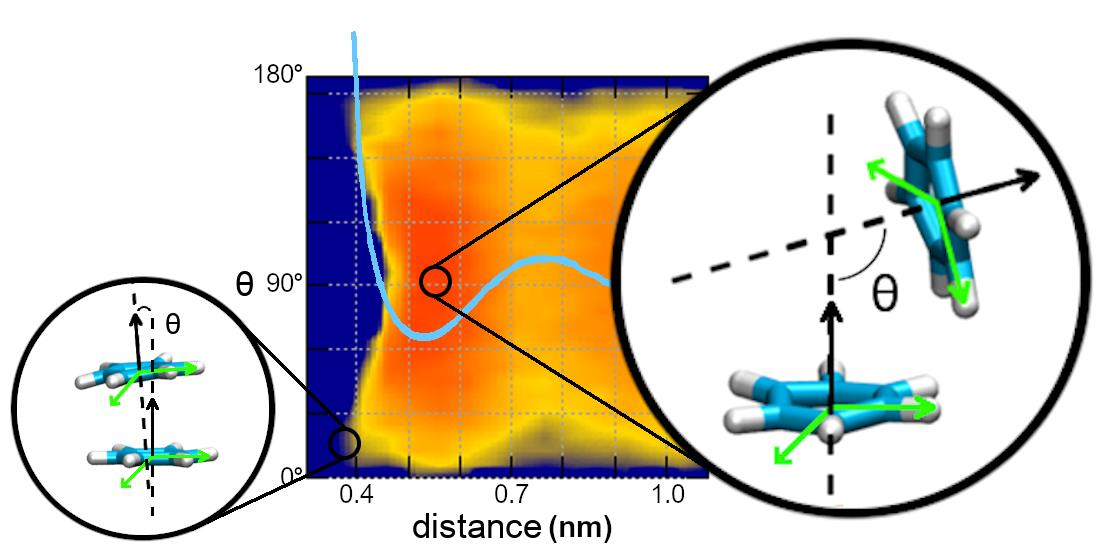
Molecules | Free Full-Text | Comparing Dimerization Free Energies and Binding Modes of Small Aromatic Molecules with Different Force Fields
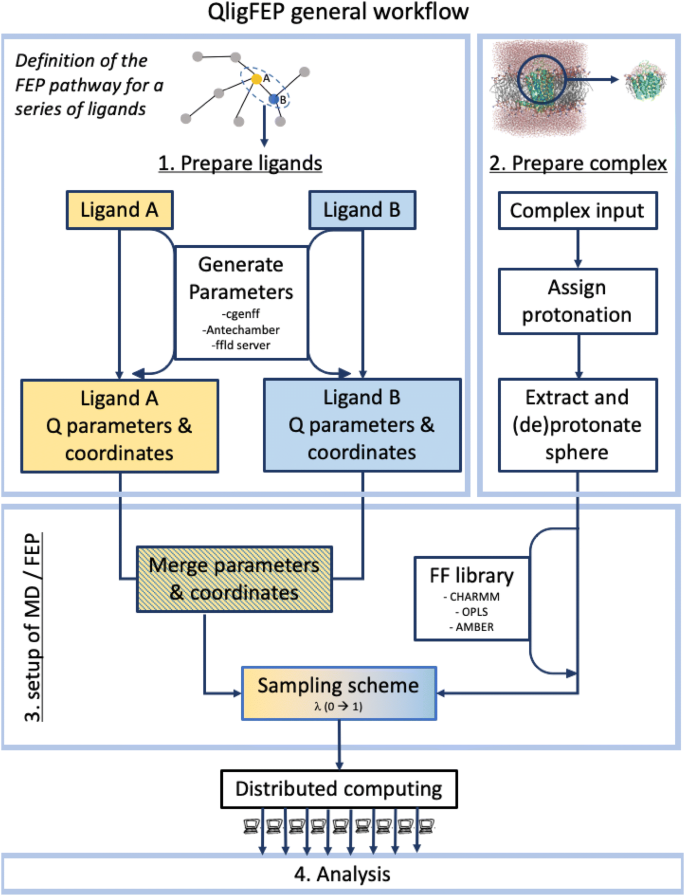
QligFEP: an automated workflow for small molecule free energy calculations in Q | Journal of Cheminformatics | Full Text

Additive CHARMM force field for naturally occurring modified ribonucleotides - Xu - 2016 - Journal of Computational Chemistry - Wiley Online Library
CHARMM force field parameters for 2'-hydroxybiphenyl-2-sulfinate, 2-hydroxybiphenyl, and related analogs

Automation of the CHARMM General Force Field (CGenFF) I: Bond Perception and Atom Typing | Semantic Scholar

Validation of the Generalized Force Fields GAFF, CGenFF, OPLS-AA, and PRODRGFF by Testing Against Experimental Osmotic Coefficient Data for Small Drug-Like Molecules | Journal of Chemical Information and Modeling
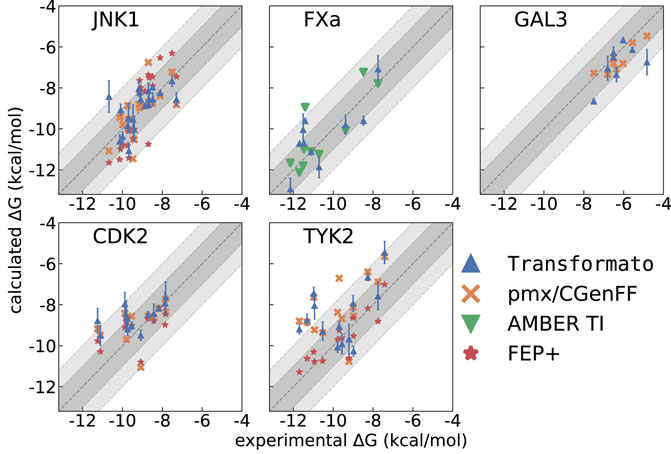
Frontiers | Relative binding free energy calculations with transformato: A molecular dynamics engine-independent tool

Validation of the Generalized Force Fields GAFF, CGenFF, OPLS-AA, and PRODRGFF by Testing Against Experimental Osmotic Coefficient Data for Small Drug-Like Molecules | Journal of Chemical Information and Modeling